
Genomic observatories
Under the genomic observatories, we investigate a range of habitats and taxa in the Belgian Part of the North Sea using DNA-based techniques. In comparison to traditional morphological analysis, DNA-based techniques allow faster screening of samples without the need for in-depth taxonomic expertise, and in some cases have proven to be more cost-efficient as well. Genomic observatories offer opportunities for the early detection of non-indigenous species, filling gaps in current regional observations and have a high level of standardisation across regions.
Genomic observatories

Traditional morphological monitoring is time-consuming, expensive and requires taxonomic experts who can be hard to find. The use of DNA-based techniques allows for faster screening of samples, without the need for in-depth taxonomic knowledge and can be standardised across regions. In many cases, the method has proven to be more time- and cost-efficient than traditional analysis, allowing for a larger spatial and temporal coverage of monitoring.
Typically, two types of DNA-based methods are used in monitoring. Current monitoring largely uses metabarcoding of bulk samples, samples containing the groups of interest. A more novel technique makes use of eDNA, or DNA that can be extracted from environmental samples like water or sediment samples.
This includes two types of DNA; 1. DNA from microscopic taxa present in the water or sediment samples themselves and 2. extra-organismal DNA originating from e.g. shed skin/tissues, excretion products, cell lysis or biologically active propagules. The latter allows us to detect the presence of a species without a specimen of the species being in the sample itself, in essence it is detected through the environment.
DNA-based methods allow us to cover a range of different taxa, including rare, cryptic or elusive taxa that are otherwise sometimes missed by traditional sampling and analysis. eDNA analysis also allows for non-invasive sampling of sensitive and rare and/or endangered taxa without putting pressure on the population.

Infrastructure
In the Belgian Part of the North Sea (BPNS), DNA and eDNA is sampled during the monthly LifeWatch campaigns by taking water samples. The grid of stations that are visited on a monthly and seasonal basis allow for a good spatio-temporal coverage of the BPNS.
LifeWatch also facilitates various other genomic monitoring events in collaboration with international consortia and Research Infrastructures like Ocean Sampling Day (OSD), EMO BON (EMBRC), ARMS-MBON (EMBRC, AssemblePlus) and Jerico.


Collected water samples are filtered onboard and stored for further processing in line with the Ocean Sampling Day protocol for analysis of microbial and plankton communities. Sequencing is performed at a centralised facility, and lab and sampling protocols are standardised.
Under EMO BON, dedicated water column and soft sediment samples are collected bi-monthly at station 330, which was selected for its long-standing biodiversity observations. EMO BON uses heavily standardised protocols for sampling and analysis and provides centralised sequencing on the European scale. Through EMO BON we target plankton communities through water column samples and microbial, meiobenthos and macrobenthos soft sediment communities in the BPNS. Through the EMO BON network, Belgium is one of the 16 connected observatories over Europe and the Mediterranean.
ARMS-MBON enables monitoring of hard substrate fauna by deploying Autonomous Reef Monitoring Reefs (ARMS) units at various locations at the BPNS during one year long deployments. The ARMS MBON project has 20 observatories all over Europe and the Arctic and provides metabarcoding of samples to its partners targeting COI and 18S markers.

Data & Services
Related stories
Relevant news
-
Sevenfold increase in stinging weever fish observed by SeaWatch-B 
Sevenfold increase in stinging weever fish observed by SeaWatch-B
Between April and late June this year, weever fish counts were seven times higher than during the same period in 2024, according to SeaWatch-B, the citizen science project of the Flanders Marine Institute (VLIZ) under the LifeWatch Belgium Programme. -
Awardees for the 2025 WoRMS Achievement & Early Career Researcher Award known 
Awardees for the 2025 WoRMS Achievement & Early Career Researcher Award known
This year, Sammy De Grave has been honored with the WoRMS Achievement Award, and the WoRMS Early Career Researcher Award goes to Sávio Gabriel Gomes Pereira. -
Noise pollution in the North Sea 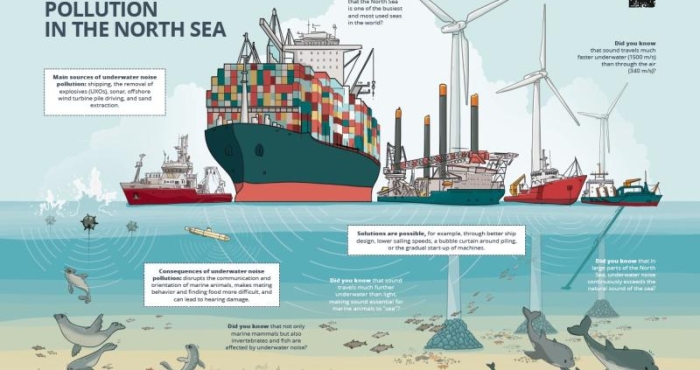
Noise pollution in the North Sea
The North Sea is one of the busiest seas in the world. All this human activity creates a lot of noise underwater. This can have harmful consequences for marine mammals and other organisms in our sea. Fortunately, measures exist to limit noise pollution. The Marine Environment Service (FOD Public Health) and DG Shipping (FOD Mobility), with the support of LifeWatch VLIZ, collaborated on a campaign to raise awareness about underwater noise pollution among the wider public. -
Old cockles and record number of shells at the eighth edition of the Big Seashell Survey Old cockles and record number of shells at the eighth edition of the Big Seashell Survey
The eighth edition of the Big Seashell Survey was a record-breaking event. No fewer than 3,500 participants scoured 400 km of coastline in northern France, Belgium, and the Netherlands, collecting and identifying nearly 150,000 shells from 72 different species. This year, the differences between the three countries were less pronounced, with the cut trough shell ranking as the most frequently found species across all regions. The Atlantic jackknife clam, the banded wedge shell, and the common cockle consistently appeared in the top five. A one-time focus on the cockle in Belgium revealed that most of these shells originated in a distant past, with only a handful of live populations remaining in the shallow continental North Sea.





