Belgian LifeWatch Observatory
Since 2012, plankton samples are collected during multidisciplinary sampling campaigns onboard of the RV Simon Stevin, visiting nine nearshore stations with monthly frequency and an additional eight offshore stations on a seasonal basis in the BPNS. As part of the Belgian LifeWatch E-science infrastructure, users interested in biodiversity and ecosystem research will be able to visualize and use the plankton data, products and web services related to plankton indicator calculations. The use case described here, has its origin in the Belgian LifeWatch Observatory.
Data system
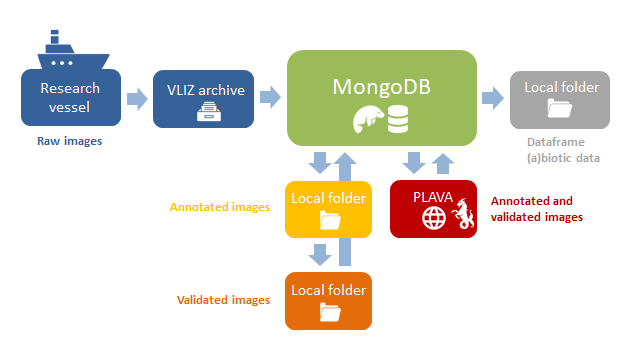
To cope and easily handle such large image datasets, both images and associated metadata are stored in BioSenseMongoDB, a NoSQL database. It is well-suited for storing image data due to its flexible and scalable document-based data model. It can handle large amounts of unstructured data, including binary image data, without the need to define a schema beforehand. Additionally, MongoDB's efficient storage and retrieval capabilities, as well as its built-in support for handling gridFS (a specification for storing and retrieving files that exceed the BSON-document size limit of 16 MB), make it an interesting choice for image data storage. The built in gridFS view is handy to look at the images directly. To date, 6.087.636 images were collected by the VPR and are stored in BioSenseMongoDB, each image associated with 28 fields containing sample-based information, 7 fields of image-associated data, 6 fields of classification data and 5 fields containing environmental-information.
In-house VPR python package and Symfony/PHP user interface (MongoDB Uploader Tool, MUT) were developed to allow scientists to easily upload raw data from the VLIZ archive to BioSenseMongoDB. Image recognition algorithms are then run on image data in BioSenseMongoDB, which will predict the taxonomic name for each image (which is important to scientists). Moreover, an in-house Graphical User Interface, the Plankton Validation Tool (PlaVa), was developed to validate collected image data, by directly querying and writing to the BioSenseMongoDB database. It allows to easily query the BioSenseMongoDB database and directly writes the new (manually assigned) validations to the database, without the need for coding or downloading the images locally.
Finally, users can query BioSenseMongoDB and extract validated biodiversity data for downstream pipelines. Image information can be reviewed and managed for quality control and IT-developments.